Aggregation Prone Regions Influences in Denaturation of Quorum Sensing Autoinducer-2 import ATP-binding Protein from Salmonella typhi Princy Vijayababu*, Gopinath Samykannu, Sundarabaalaji Narayanan
Structural Biology Laboratory, Department of Bioinformatics, Bharathiar University, Coimbatore, Tamil Nadu, India
*Corresponding author: Princy Vijayababu
Citation: Vijayababu P, Samykannu G, Narayanan S (2020) Aggregation Prone Regions (APRs) Influence in Denaturation of Quorum Sensing Autoinducer 2 import ATP-binding Protein from Salmonella typhi. Sci Academique 2(1): 01-19.
Received date: 14 December, 2020; Accepted date: 13 January 2021; Publication date: 16 January 2021
Abstract
Quorum Sensing (QS) is a cell-to-cell communication mechanism in bacterial system for harmonizing group behaviors such as developing biofilms and producing virulence factors. In Salmonella typhi, Autoinducer (AI-2) is a small signaling molecule for interspecies communication, LsrA is an ATP-binding membrane protein responsible for energy coupling in ABC transport of quorum sensing system. Obtaining biologically active membrane protein has been a challenge since the poor solubilization yield. Reducing the protein aggregation is the only straight forward task to increase the yield of protein. The aggregation behavior of a protein is strongly determined by intrinsic properties of its amino acid sequence and mediated by short Aggregation Prone Regions (APRs). Denaturation of Autoinducer-2 import ATP-binding protein from S. typhi, using an additive is the main objective of this study. Aspartic acid used as the additive has been found to reduce the number of APRs as structural gatekeeper. This is an interdisciplinary study to improve the yield of Salmonella typhi LsrA, and can be used for further structure and functional characterization studies in various field.
Keywords: Quorum Sensing; ABC transporter; Aggregation Prone Regions; Structural gatekeeper
Introduction
Gram -negative bacteria including Salmonella typhi, talk to one another by “Quorum Sensing” (QS), which allow the bacterial group to coordinate gene expression with the population of cell density, which can harmonize group behaviors such as forming biofilms and producing virulence factors. [1,2]. This intercellular communication is accomplished through the production, exchange and release of small chemical signaling molecules called autoinducers (AI) [1]. Auto inducers are made from S- adenosylmethionine, and used as a methyl donor in a variety of cellular processes which yield S-adenosylhomocysteine. S-Adenosylhomocysteine is consequently metabolized to adenine and S-ribosylhomocysteine. S- Ribosylhomocysteine is the substrate for LuxS, which cleaves it to produce homocysteine and 4, 5-dihydroxy-2, 3-pentanedione (DPD). DPD cyclizes spontaneously and undergoes further rearrangements to form AI-
2. It is internalized by the Lsr ABC-type transporter. LsrA is an ATP- binding membrane protein responsible for energy coupling in ABC transport of quorum sensing system [3].
Obtaining biologically active membrane protein has been a challenge since the poor solubilization yield [4,5]. In a majority of cases, the overexpressed protein accumulates intracellularly forming a dense amorphous insoluble aggregate known as inclusion bodies [6]. To prepare very pure inclusion bodies, characterization studies, washing with a buffer containing detergent and additives will help to remove protein aggregation [7]. Indeed, reducing the protein aggregation is the only straight forward task to increase the yield of protein [8]. The aggregation behavior of a protein is strongly determined by intrinsic properties of its amino acid sequence [9]. This aggregation mediated by short Aggregation Prone Regions (APRs) [10].
In this background our attempts have been made to denature the Autoinducer 2 import ATP-binding protein from S. typhi, by using an additive, which has been found to reduce the number of APRs as structural gatekeeper. This is the interdisciplinary study to improve the yield of Salmonella typhi LsrA, and can be used for further structure functional characterization studies in various field.
Materials and Methods
Molecular Cloning
lsrA gene was amplified with gene specific primers (Forward primer: 5’GCGCATATGCAAATCAGTCACAATACTG 3’ and Reverse primer:
5’GCGCTCGAGACATGTTGCCTCCGACG 3’ containing NdeI and Xh oI restriction sites, respectively) using the S. typhi Ty21a genomic DNA as the template by PCR (MULTIGENE OPTIMAX). Amplified gene product was digested with NdeI and XhoI (New England Biolabs) sites and ligated into pET30b (+) vector (Novagen) for overexpression. The ligation mixture was transformed into chemically competent E. coli DH10β cells by CaCl2 heat-shock method and plated on LB- kanamycin (30 µg/ml) followed by incubation at 37°C for 12 hrs. Transformed colonies were screened by colony PCR using gene specific primers. Plasmid DNA was isolated and verified for presence of insert using restriction digestion with NdeI and XhoI sites.
Overexpression of LsrA
The confirmed recombinant plasmids lsrA– pET30b (+) were transformed into competent E. coli BL21 (DE3) expression host strain. From the transformed colonies cells were selectively picked and subsequently transferred into a 50 mL of LB medium supplemented with kanamycin (30 µg/mL). The bacterial cells were then allowed to grow inside an orbital shaker incubator at 160rpm/mins at 37 °C. When the OD600nm of each flask was reached, the protein production was induced by the addition of different concentrations of IPTG (0.1, 0.2 and 0.4 mM) and grown at 37°C for 4hrs. For the temperature optimization, the bacterial culture was induced with 0.1mM IPTG and cultured at three different temperatures (4, 20 and 37 °C) with 3,6 and 12hrs incubation time interval. The sample from each optimization experiment was collected and the absorbance at OD600nmwas determined in order to monitor the bacterial growth. The cell cultures exhibiting maximum growth were harvested and centrifuged (SORVALL-ST16R) at 10,000 rpm for 10 mins at 4 °C following which all the fractions (supernatant and pellet) were analyzed by 12%SDS– PAGE.
Optimization of solubilization using various agents
Using different concentrations of IPTG (0.1, 0.2 and 0.4 mM), varying the temperature (4, 20 and 37 °C), incubation time and host cells BL21(DE3), LB Media (Luria-Bertani broth-LB), additive and different molar concentration of chaotropic agents’ attempts were made to express the S. typhi LsrA in the soluble form. Nonetheless, all the optimization steps have resulted in the expression of recombinant LsrA as inclusion bodies only (pellet fraction).
In silico identification of aggregation prone regions (APRs)
The primary sequence of S. typhi LsrA comprising 511 amino acid residues were retrieved from UniProtKB (Q8Z2X5) [11] and were subjected to ProGene1.0 [12] for calculating molecular weight and theoretical Iso electric point (PI). AGGRESCAN server is a web-based application used to predict APRs. The aggregation-propensity value for each residue in the LsrA sequence was computed by averaging aggregation-propensity value per each amino acid [13].
Identification of Structural Gatekeepers
AGGRESCAN server calculates the aggregation propensity of each single amino acids according to its relative position. However, in order to cut down all the APRs and to know the best structural gatekeepers, 20 amino acids were replaced at the central hydrophobic cluster in each APRs.
Denaturation of LsrA with additives
Purified IBs were harvested and resuspended in 30 mL of solubilization buffer containing (8M Urea) and were further diluted into buffer containing 50mM Tris-Cl (pH8.0),100mM NaCl, 2% Triton X-100, 5mM DTT and 10MmAspartic acid as additive and allowed to solubilize with moderate shaking at 37°C for overnight [14]. Solubilized LsrA was centrifuged at 15,000 rpm for 90 mins at 20 °C. The fractions (Supernatant and pellet) were examined by 12% SDS-PAGE. Over all methodology were shown in Figure 1.
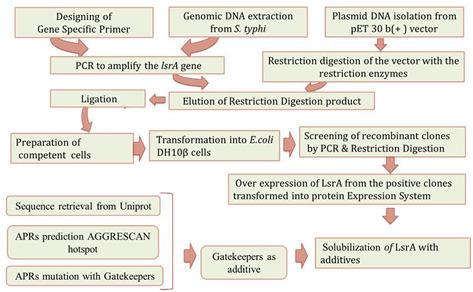
Figure 1: Overview of LsrA Denaturation with APRs. Direction of arrow indicate the work direction start from Gene specific primer designing and end with LsrA solubility with additive.
Results
Recombinant LsrA: lsrA gene was cloned between the NdeI and XhoI sites of prokaryotic expression vector pET30b(+). Double digestions were conducted to examine, the amplified product of lsrA gene from S. typhi and recombinant plasmid (pET30b(+)-lsrA). As shown in supplementary figure 1, the size of target fragment from double digestion of pET30b (+)-lsrA released at the position of 1533 bp (supplementary figure 2).
Overexpression of LsrA
The cloned plasmid was transformed into optimized expression host (E. coli BL21 (DE3)) cells. Expression of the recombinant construct was under the control of a T7 promoter, which can be easily induced for expression by isopropyl β-D-1-thiogalactopyranoside (IPTG). Optimized condition of 0.1mM IPTG induction has revealed an increased presence of a band with an approximate molecular weight of 55kDa, S. typhi LsrA induced and uninduced samples have been shown in Figure 2.
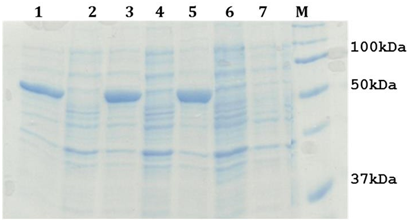
Figure 2: 12% SDS-PAGE analysis of LsrA over production. Lane 1, 3 and 5: A cell free extract of E. coli BL21 (DE3) containing pET30b(+)-lsrA induced sample, Lane 2 and 4: A cell free extract of E. coli BL21(DE3) containing pET30b(+)-lsrA uninduced sample, Lane 6 and 7: A cell free extract of E. coli BL21(DE3) containing pET30b (+) induced and uninduced sample, Lane M : Protein Molecular Weight Marker.
Extraction and purification of LsrA Inclusion Body (IBs): In order to check the existence of S. typhi LsrA in pellet (inclusion bodies), the cell pellet of induced culture was suspended in 50 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1mM PMSF, 1% Tween 20 and 70 µg/ml Lysozyme without any denaturant. Cells were disrupted by ultra-sonic waves generated using
SONICS, 130W using an optimized run time of 30mins with a pulse of 3 seconds on and 6 seconds off on ice. Crude inclusion bodies were obtained by centrifugation of the cell lysate at 10,000 g for 10 mins at 4 °C and were analyzed on12% SDS-PAGE. After confirming the presence of inclusion bodies in the pellet, they were washed thrice in 10 mL (1:10) of lysis buffer. Following this, the pellet was subjected for extensive final wash with buffer A in without 1% Triton X-100 and 2M Urea. These re- suspended inclusion bodies were kept at 37 °C on incubator shaker for 15 mins and centrifuged (7,000rpm, 7 mins at 4 °C) to remove soluble and contaminating proteins. The supernatant and pellet obtained during every stage of the IBs purification process were analyzed by 12% SDS PAGE.
Denaturation of LsrA
Purified LsrA inclusion bodies were done in various denaturation buffers with different concentrations of choatropic agents, different temperatures and pH. Solubilization was done for 12 hrs at 37 °C with moderate shaking. Undissolved inclusion bodies were removed by centrifugation at 13,000g for 45 min at 4 °C to obtain solubilization LsrK; remains the pellet not in the Figure 3. Screen with different buffers, pH, different concentration of urea, reducing agent and Imidazole were tried out as shown in the table 1. In this manner we introduced in silico approach to predict aggregation prone regions(APRs).
|
Tris-Cl /pH |
NaCl |
PMSF |
Lysozyme |
Reducing agents |
Imidazole |
EDTA |
Detergents |
Chaotropic agent |
|
100mM/8 |
500mM |
1 mM |
– |
1% β-ME |
10 mM |
10 mM |
2% Triton X-100 ,2% SDS |
– |
|
100mM /8 |
500mM |
1 mM |
20 µg/ml |
1% β-ME |
10 mM |
10 mM |
2% Triton X-100 , 2% Tween 20,2% SDS |
– |
|
50mM/8 |
500mM |
1 mM |
– |
3% β-ME |
10 mM |
10 mM |
2% Triton X-100 ,2% Tween 20 |
– |
|
100mM/8 |
500mM |
1 mM |
– |
1% β-ME |
– |
10 mM |
2% Triton X-100 |
8 M Urea |
|
100mM/8 |
500mM |
1 mM |
– |
3mM DTT |
– |
10 mM |
2% Triton X-100, 3mM DDM |
8 M Urea |
|
50mM/8 |
50mM |
1 mM |
– |
3mM DTT |
– |
20 mM |
2% Triton X-100 , 3mM DDM |
8 M Urea |
|
50mM/8 |
500mM |
1 mM |
– |
3mM β-ME |
– |
– |
2% Triton X-100 |
– |
|
50mM/8 |
500mM |
1 mM |
– |
3mM β-ME |
– |
– |
2% Triton X-100 |
8 M Urea |
|
50mM/8 |
500mM |
1 mM |
– |
3mM β-ME |
– |
– |
2% Triton X-100 |
8 M Urea |
|
100mM/12 |
500mM |
1 mM |
70 µg/ml |
3mM β-ME |
– |
20 mM |
2% Triton X-100 |
8 M Urea |
|
50mM/10 |
500mM |
1 mM |
– |
3mM β-ME |
– |
– |
2% Triton X-100 |
8 M Urea |
|
50mM/8 |
50mM |
1 mM |
70 µg/ml |
3mM DTT |
– |
20 mM |
2% Triton X-100 |
8 M Urea |
|
50mM/8 |
50mM |
1 mM |
70 µg/ml |
3mM, 3mM DTT |
– |
20 mM |
2% Triton X-100 |
8 M Urea |
|
Salt concentration were varied from 50 mM-100 m. Lysozyme, Reducing agents, imidazole and choatropic agent (Urea) were used in different concentration. Nevertheless S. typhiLsrA remain in pellet fraction because of protein aggregation. |
||||||||
Table 1: Optimization of LsrA solubilization buffer.
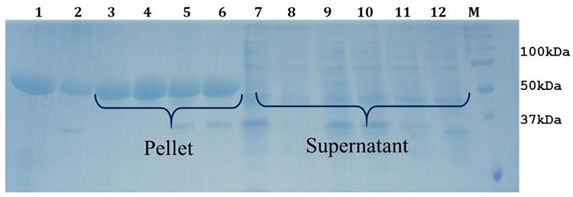
Figure 3: 12% SDS-PAGE analysis of LsrA solubilization using Urea.Lane1: LsrA whole cell pellet. Lane 2: whole cell lysate. Lane 3 and 7: LsrA solubilization of supernatant and pellet with Tris-Cl (pH 12.0), Lane 4 & 8: LsrA solubilization of supernatant and pellet with Tris-Cl (pH 10.0), Lane 5 & 9: LsrA solubilization of supernatant and pellet using 500 mM NaCl, Lane 11: LsrA solubilization of supernatant using 70µg/ml lysozyme, Lane M: Protein Molecular Weight Marker.
In silico identification of APRs
The prediction of APRs is based on the primary sequence of a protein [15]; accordingly, we carried out the primary sequence investigation of S. typhi LsrA, 511 amino acid residues of LsrA were analyzed using ProGene1.0, which shows the molecular weight and isoelectric point as 55.8 kDa and 5.9 respectively. The results of AGGRESCAN 13 server have revealed the presence of 22 APRs which are shown in table 2.
|
S.No |
Start position |
Hot spot Peptide |
End position |
|
1. |
11 |
LICVQNIYK |
19 |
|
2. |
21 |
YSGVEVLKGIDF |
32 |
|
3. |
52 |
TLMKIIAGIV |
61 |
|
4. |
70 |
IAGVRCSHL |
78 |
|
5. |
83 |
AHQYGIYLV |
91 |
|
6. |
100 |
SLSVRENIL |
108 |
|
7. |
125 |
LKAMG |
129 |
|
8. |
150 |
EIMRG |
154 |
|
9. |
156 |
MRDSR |
160 |
|
10. |
188 |
KKGVGIVFISHKL |
200 |
|
11. |
205 |
QLAHCVS |
211 |
|
12. |
213 |
MRDGK |
217 |
|
13. |
231 |
EIIQAIT |
237 |
|
14. |
247 |
ASQKLWL |
253 |
|
15. |
269 |
LALESLT |
275 |
|
16. |
289 |
GEILGLAGLVGAG |
301 |
|
17. |
307 |
ETLYGIR |
313 |
|
18. |
336 |
LQLGLVYL |
343 |
|
19. |
353 |
YLDASLAWNVC |
363 |
|
20. |
413 |
LIAKCLEAS |
421 |
|
21. |
450 |
AQQNVAVLFISSD |
462 |
|
22. |
494 |
DTIMHVAF |
501 |
|
AGGRESCAN server is predict 22 APRs (Hotspot peptide) with hydrophobic centralized position, which have aggregation propensity. |
|||
Table 2: Identification of APRs from LsrA.
In order to eliminate the APRs amino acid residues forming the hydrophobic cores Q15, L27, I57, R74 , G87, R104, A127, M152,158D,V194, H208, D215, Q234, K250, E272, A295,Y310, L340, L358, C417, V456 and H498 of LsrA were initially identified. But most often they form an integral part of the hydrophobic core of the protein [16,17] and protected from higher-order interactions. Mutating a central hydrophobic residue in the APRs with twenty amino acids and reduced the number of APRs [16] and total area of aggregation has been shown in Figure 4.
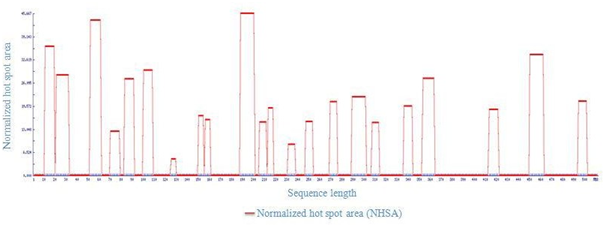
Figure 4: Total Area of host spot from LsrA. 22 hostspot share 44.789 from total area of 4.89.
Point mutation with structural gatekeepers and their amino acid aggregation-propensity values such as Sequence Average, Number of Hot Spots, Normalized number of Hot Spots for 100 residues, Total Area and other details of mutation were shown in Table 3. From the overhead APRs analysis Q15P, L27D, I57P, R74D, G87P, R104P, A127D, M152D,V194P, H208D, D215P, Q234D, K250D, E272P, A295D,Y310D, L340D, L358D, C417D, V456D and H498P mutations were selected as the enhanced point mutation in LsrA. As the result of these point mutations, hotspots become null hot spot as shown in supplementary Figure 3-5. From these results, aspartic acid has been found to be the best structural gatekeeper for S. typhi LsrA.
|
Protein Sequence Name |
LsrA |
|
A3v Sequence Average (A3VSA) |
-0.006 |
|
Number of Hot Spots (NHS) |
22 |
|
Normalized NHS For 100 Residues (NNHS) |
4.305 |
|
Area of the Profile Above Threshold (AAT) |
53.95 |
|
Total Hot Spot Area (THSA) |
44.783 |
|
Total Area (TA) |
4.89 |
|
Aat Per Residue (AATR) |
0.106 |
|
Thsa Per Residue (THSAR) |
0.088 |
|
Normalized A4v Sequence Sum For 100 Residues (NA4VSS) |
-1.1 |
Table 3: Aggregation-propensity value for S, typhi LsrA.
Solubilization of LsrA using additives (Aspartic acid)
Structural gatekeeper of aspartic acid was introduced as additive in LsrA solubilization buffer composition (50mM Tris-Cl (pH8.0),150mM NaCl, 8M Urea, 1% Triton X-100,3mM β-ME and 10 mM aspartic acid). Finally, the LsrA solubilization was standardized with 8M Urea and 10 mM aspartic acid. This revealed, Urea as the strong denaturing agent and aspartic acid as a good additive, that has the ability to solubilize LsrA and other additives (Figure 5). Removal of aspartic acid from the purified protein is also easier due to its molecular weight and has lower effect on protein solubilization as compared to other additives [14]. This standardized buffer could be used in further experiments for solubilization of LsrA.
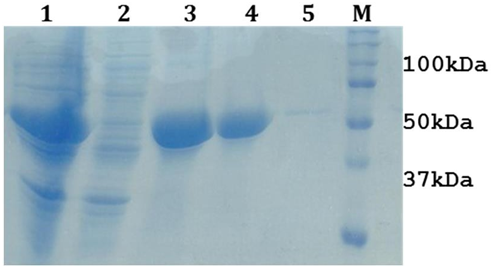
Figure 5: 12% SDS-PAGE showing the optimization of LsrA solubilization. Lane1 & 2 represents LsrA whole cell pellet & lysate (after sonication), Lane 3 & 4: LsrA-IBs, Lane 4: LsrA solubilization in supernatant (dissolved in 8M Urea with 10mM aspartic acid) Lane 5: LsrA solubilization in pellet, Lane M: Protein Molecular weight marker .
Optimization of LsrA IBs
The absorption maxima at wavelength 280 (OD280) of the inclusion bodies solubilized in the Tris-urea buffer ranging from 2 M to 8 M urea showed maximum absorption for the sample solubilized in 8 M urea (Table 4). This indicates that maximum solubilization occurred in 8 M urea. Figure 6 showed that absorption maxima increased sharply from 4 M to 6 M urea solubilization and saturation in solubilization till 8 M urea. Therefore OMPLA inclusion bodies were denatured in 6 to 8 M urea for achieving maximum solubilization.
|
Molarity |
OD 280 |
|
1M |
0.328 |
|
2M |
0.488 |
|
3M |
0.666 |
|
4M |
0.925 |
|
5M |
0.969 |
|
6M |
0.945 |
|
7M |
0.923 |
|
8M |
0.902 |
Table 4: Screen for optimizing conditions for solubilization of LsrA from inclusion bodies using UV spectrophotometer.
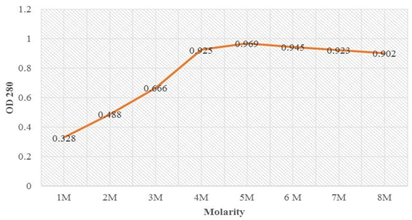
Figure 6: Optimization denaturation of LsrA using different urea concentration showing sharp rise in the absorption from 4M to 6M urea concentration and further on stabilized till 8M urea denaturation.
Discussion
Membrane proteins contain large hydrophobic patches on their surfaces that enable the proteins to occupy the hydrophobic interior of a lipid bilayer in a membrane [18]. Size and density of the inclusion bodies depend upon the expression level and nature of protein. To prepare very pure inclusion bodies for subsequent characterization studies, washing with a buffer containing detergent and additives will most likely take away protein aggregation [7]. Accordingly, we optimize the solubilization buffer with different additives, pH, and salt concentrations.
Protein aggregation is very difficult to remove without affecting protein stability and structure. Besides structural stabilization, a number of mechanisms can contribute to reduce aggregation [19]. Usually charged amino acids (Arg, Lys, Glu and Asp) or proline, are predominantly used as structural gate keepers and they need to be placed in loop regions in order to stabilize the structure [17]. In general, the solubility of proteins is directly related to the number of APRs found within a protein sequence. Over 80% of proteins possess at least one APR in their primary sequence [15], but most often they form an integral part of the hydrophobic core of the protein [16,17] and protected from higher-order interactions. Fascinatingly, recent algorithm (AGGRESCAN) takes not only the primary sequence of the polypeptide, but also experimental proteome data and the aggregation propensity of bacterial proteins associated with their length, conformation, location, function and abundance [13]. Our other proteomics studies show the importance of protein structure and characteristics [20-23]. These reported studies have given the significant support to incorporate our experimental results to in silico approach.
Conclusion
S. typhi lsrA gene was cloned into pET30b (+) vector and overexpressed under control of the T7 promoter using E. coli BL21 (DE3) cells. To solubilize LsrA inclusion bodies with Host strains, Temperature, Incubation period, pH, Choatrops, Detergents, Additives and Solubilization buffers (Tris-Cl) were optimized gradually. The objective was then focused to In-silicopart in order to acquire aggregation prone regions. In our study, 22 APRs were revealed by AGGRESCAN server and successful point mutations of Q15P, L27D, I57P, R74D, G87P, R104P, A127D, M152D, V194P, H208D, D215P, Q234D, K250D, E272P, A295D, Y310D, L340D, L358D, C417D, V456D and H498P were carried out. Aspartic acid is a reminiscent of aggregation gatekeeper and used as an additive for LsrA denaturation. As evident of our study, clearly shows that addition of aspartic acid assists in denaturation of S. typhi LsrA.
Acknowledgement: The authors thank to I. Karthik, CAS in Crystallography and Biophysics, University of Madras, Chennai-India, for valuable suggestion in the production of the manuscript.
Declaration of interests: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Xavier KB, Bassler BL (2005) Interference with AI-2-Mediated Bacterial Cell-Cell Communication. Nature 437: 750–753.
- Vijayababu P, Samykannu G, Narayanan S (2017) Structural insights of cell-cell communication autoinducer-2 kinase (LsrK) model from Salmonella typhi. International Journal of Current Research 9: 51169- 51172.
- Xue T, Zhao L, Sun H, Zhou X, Sun B (2009) LsrR-binding site recognition and regulatory characteristics in Escherichia coli AI-2 quorum sensing. Cell Research 19: 1258-1268.
- Singh A, Upadhyay V, Upadhyay AK, Singh SM, Panda AK (2015) Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microbial cell factories 14: 41.
- Vijayababu P, Samykannu G, Antonyraj CB, Thomas J, Narayanan S, et al. (2018) Patulin interference with ATP binding cassette transferring auto inducer −2 in Salmonella typhiand biofilm inhibition via quorum sensing. Informatics in Medicine Unlocked 11: 9–14.
- Kopito RR (2000) Aggresomes, inclusion bodies and protein aggregation. Trends in Cell Biology, 10: 524–530.
- Singh A, Upadhyay V, Panda AK (2014) Solubilization and Refolding of Inclusion Body Proteins. Insoluble Proteins: 283–291.
- De Marco A, Vigh L, Diamant S, Goloubinoff P (2005) Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol-overexpressed molecular chaperones. Cell stress & chaperones 10: 329-39.
- Castillo V, Graña-Montes R, Sabate R, Ventura S (2011) Prediction of the aggregation propensity of proteins from the primary sequence: Aggregation properties of proteomes. Biotechnology Journal 6: 674– 685.
- Beerten J, Schymkowitz J, Rousseau F (2013) Aggregation Prone Regions and Gatekeeping Residues in Protein Sequences. Current Topics in Medicinal Chemistry 999: 43–49.
- Apweiler R, Bairoch A, Wu CH (2004) Protein sequence databases. Current Opinion in Chemical Biology 8: 76-80.
- Vijayababu P, Samykannu G, Narayanan S (2017) ProGene 1.0-An In silico Tool for Protein-Gene Analysis. International Journal of Life- Sciences Scientific Research 3: 1089-1093.
- Conchillo-Solé O, de Groot NS, Avilés FX, Vendrell J, Daura X, et al. (2007) AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics 8: 65.
- Samykannu G, Vijayababu P, Antonyraj CB, Narayanan S (2020) Structural investigation of APRs to improve the solubility of outer membrane protease (PgtE) from Salmonella enterica serotype typhi- A multi-constraint approach. Biochemistry and Biophysics Reports 21: 100693.
- Rousseau F, Schymkowitz J, Serrano L (2006) Protein aggregation and amyloidosis: confusion of the kinds? Current Opinion in Structural Biology 16: 118–126.
- De Baets G, Schymkowitz J, Rousseau F (2014) Predicting aggregation-prone sequences in proteins. Essays In Biochemistry 56: 41–52.
- Ganesan A, Siekierska A, Beerten J, Brams M, Van Durme J, et al. (2016) Structural hot spots for the solubility of globular proteins. Nature Communications 7: 10816.
- Lin JH, Baumgaertner A (2000) Molecular dynamics simulations of hydrophobic and amphiphatic proteins interacting with a lipid bilayer membrane. Computational and Theoretical Polymer Science 10: 97– 102.
- Stefani M, Rigacci S (2013) Protein Folding and Aggregation into Amyloid: The Interference by Natural Phenolic Compounds. International Journal of Molecular Sciences 14: 12411–12457.
- Samykannu G, Vijayababu P, Antonyraj CB, Perumal P, Narayanan S, et al. (2019) In Silico Characterization of B Cell and T Cell Epitopes for Subunit Vaccine Design of Salmonella typhi PgtE: A Molecular Dynamics Simulation Approach. Journal of Computational Biology 26: 105–116.
- Vijayababu P, Samykannu G, Antonyraj CB, Narayanan S, Basheer Ahamed SI, et al. (2018) B-cell and T-cell epitope identification with stability analysis of AI-2 import ATP-binding cassette LsrA from S. typhi In silico approach. Microbial Pathogenesis 123: 487–495.
- Samykannu G, Vijayababu P, Natarajan J (2018) Substrate specificities in Salmonella typhi outer membrane protease ( PgtE ) from Omptin family – An in silico proteomic approach. Informatics in Medicine Unlocked 12: 6–13.
- Samykannu G, Vijayababu P, Antonyraj CB, Narayanan S, Basheer Ahamed SI (2017) Investigations of binding mode insight in Salmonella typhi type-III secretion system tip protein (SipD): A molecular docking and MD simulation study. Informatics in Medicine Unlocked 9: 166–172.
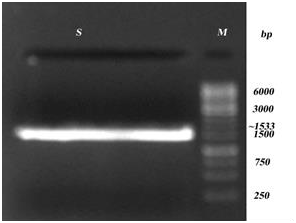
Supplementary Figure 1: PCR amplification of S. typhi- lsrK. Lane S: Amplified lsrK, Lane M- Marker.
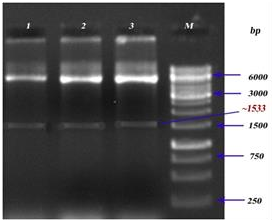
Supplementary Figure 2: Restriction digestion analysis of LsrA. Lanes 1, 2&3: Recombinant construct of lsrA (pET30b(+)-lsrA), Lane M:Marker.
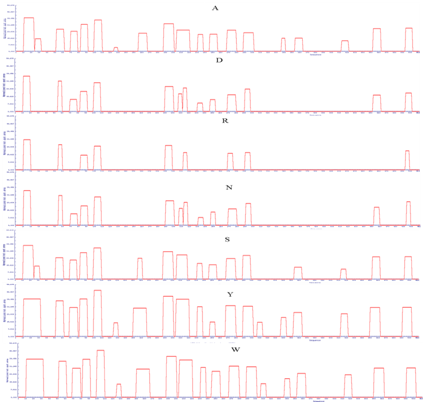
Supplementary Figure 3: APRs area of LsrA. Amino acid indicate the mutated amino acid propensity area.
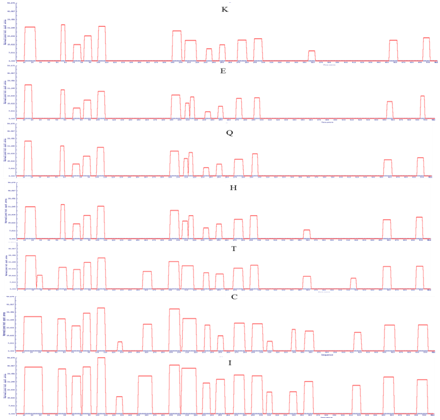
Supplementary Figure 4: APRs area of LsrA. Amino acid indicate the mutated amino acid propensity area.
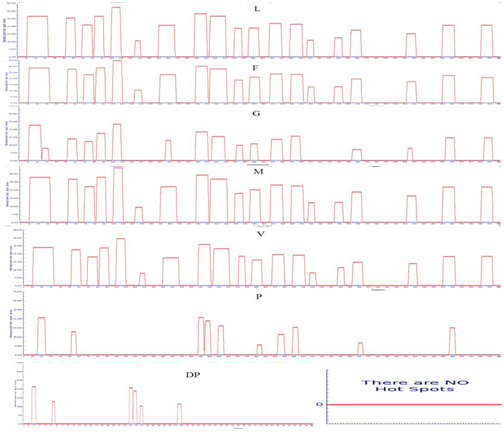
Supplementary Figure 5: APRs area of LsrA. . Amino acid indicate the mutated amino acid propensity area.


