rs1801282 (Pro12Ala) single nucleotide polymorphism in the PPARG gene is associated with gestational diabetes mellitus among Filipinos Maria Ruth Pineda-Cortel1,2*, Sheena Abad 1, Yvette Bailon1, Vanessa Mae Bugarin1, Ryna Zatchel Domingo1, John Lawrence Fernandez1, Trisha Claire Mendoza 1, Raphael Enrique Tiongco3
1Department of Medical Technology, Faculty of Pharmacy, University of Santo Tomas, Manila, Philippines
2Research Center for the Natural and Applied Sciences, University of Santo Tomas, Manila, Philippines
3Department of Medical Technology, College of Allied Medical Professions, Angeles University Foundation, Angeles City, Philippines
*Correspondence to: Maria Ruth Pineda-Cortel
Citation: Pineda-CortelMR, AbadS, BailonY, Bugarin VM, Domingo RZ, et al. (2023) rs1801282 (Pro12Ala) single nucleotide polymorphism in the PPARG gene is associated with gestational diabetes mellitus among Filipinos. Sci Academique 4(1): 43-54
Received: 12 May, 2023; Accepted: 05 June 2023; Publication: 09 June 2023
Abstract
Aims: In this study, we determined the association between rs1801282 (Pro12Ala), a genetic variant of the PPARG gene, with GDM among Filipinos.
Methods: A cross-sectional design study was utilized, and with ethical approval, 258 pregnant women (86 cases and 172 controls) from various clinics and hospitals in Metro Manila, Philippines were recruited. Whole blood and serum samples were taken from each participant for analysis. Laboratory tests such as oral glucose tolerance test, glycated hemoglobin, and lipid profile were performed. Genomic DNA was extracted from the buffy coat and was subjected to analysis using TaqMan assay to identify the genetic polymorphism.
Results: Results of the study suggest that the C/G and G/G genotypes are significantly associated with GDM development. Moreover, the presence of C/G and G/G genotypes are significantly associated with fasting blood sugar, total cholesterol, and low-density lipoprotein cholesterol levels. On the other hand, the C/C genotype is significantly correlated with triglycerides and very low-density lipoprotein levels.
Conclusion: Overall, a significant association between the rs1801282 polymorphism of the PPARG gene and GDM was observed. However, further studies are still needed to confirm the results of our study.
Keywords: Gestational diabetes mellitus; Single nucleotide polymorphism; rs1801282; peroxisome proliferator-activated receptor gamma; Filipino
Introduction
Gestational diabetes mellitus (GDM), one of the most recurrent metabolic diseases during pregnancy, has been tagged as “a disease across two generations,” as it affects both the mother and the baby. World Health Organization defines it as any degree of glucose intolerance with first recognition during pregnancy. Although, it disappears immediately after baby delivery, it is associated with various maternal, neonatal and fetal complications. Maternal complications include pre-eclampsia, antepartum hemorrhage, urinary tract infection. While fetal and neonatal complications include macrosomia, fetal malformation, sepsis, neural tube defects, cardiovascular defects, and among others [1,2]. Long term complications are also observed, such as the risk for metabolic syndrome, obesity, hypertension, and type 2 diabetes mellitus (T2DM) [3]. Studies have shown that mothers with GDM and babies born from mothers with GDM have 15 to 50% chance of developing T2DM [4]. Pregnant women worldwide suffer from this health condition, which is a complex disease resulting from many risk factors, such as sedentary lifestyle, age, weight, genetics, and lipoprotein levels. Several studies have been conducted that determined the association of various genetic variants with the development of GDM and even lipoprotein levels [5]. Abdominal obesity, sedentary lifestyle, advancing age, and genetic and epigenetic factors affecting glucose homeostasis are thought to be key contributing factors [6]. Numerous genetic markers have been associated with GDM, one of which that shows a promising correlation is the peroxisome proliferator-activated receptor gamma (PPARG).
The PPARG is a ligand-activated transcription factor located at 3p25, which plays an essential role in the regulation of lipid storage, fatty acid uptake, glucose uptake, and energy balance. It has a significant role in the formation of adipose tissue and the subcellular metabolism of the macrophage cells in the arterial wall [7]. The PPARG also activates genes involved in both glucose and insulin metabolism [8,9]. Several single nucleotide polymorphisms (SNPs) in the PPARG have been reported to be associated with lipoprotein levels. From among these polymorphisms, one significant SNP is the rs1801282 (Pro12Ala) in the PPARG that has been proven to be related to lipoprotein levels of certain individuals [10–12]. The rs1801282 is a polymorphism that results in the CCA to GCA or proline to alanine substitution located at the exon 2 codon 12 of the PPARG gene [13,14]. Multiple studies have stated that the Pro12Ala polymorphism improves insulin sensitivity in humans [15,16]. Individuals who have at least one copy of Ala have lower BMI and higher insulin sensitivity. Also, having the Ala allele appeared to have a protective role against T2DM [17]. However, there are also studies showing a completely different result, that is, Pro12Ala polymorphism is linked to risk of T2DM [14].
Several studies on the association of the rs1801282 SNP in the PPARG gene with the incidence of T2DM and its possible effect on serum lipoprotein levels and metabolic syndrome have been conducted. However, limited studies included pregnant women with GDM, and none in the Philippines. GDM has been said to have the same pathophysiology or hallmark with T2DM, that is, insulin resistance. Hence, in this study, we determined the association of the rs1801282 polymorphism with the incidence of GDM among pregnant women in the National Capital Region, Philippines using a cross-sectional research design. Also, we determined the association of the said polymorphism with blood glucose, glycated hemoglobin, and serum lipid profile levels among the study population.
Materials and Methods
Research design, respondents, and study site
This study used a cross-sectional design wherein the exposure (presence of the SNP) and outcome (presence of GDM) of pregnant women were simultaneously measured. A total of 258 pregnant women was recruited using a purposive sampling technique and grouped into GDM (86) and non-GDM (172) following oral glucose tolerance and physician’s diagnosis. Participants included were pregnant women aged 18 to 45 years old and are free from any recognizable diabetes. Those excluded were pregnant women diagnosed with metabolic disease (e.g., thyroid disease, polycystic ovarian syndrome) before pregnancy and those with a previous history of GDM. All pregnant women participating in the study were recruited from various hospitals, birthing clinics, and clinical laboratories in the National Capital Region, Philippines.
Patient interview, physical assessment, and sample collection
Using a structured interview guide, information such as age, address, occupation, civil status, nationality, medical, family, and obstetric history were obtained. Height, pre-gestational, and gestational weight and age of gestation were also determined. From these measured variables, body mass index (BMI) was computed. Blood samples were collected from each participant. Serum samples were obtained and were used for oral glucose tolerance test (OGTT) and lipid profile testing. Anticoagulated whole blood was used for glycated hemoglobin (HbA1c) testing and genomic DNA extraction.
Biochemical analysis
Two-hour OGTT using 75 grams glucose load was used to determine if participants have GDM or none. The routine glucose oxidase-peroxidase coupled reaction was used to quantitate the concentration of glucose. Results of the OGTT 75-grams was then interpreted using the IADPSG criteria as recommended by the results of a previous study by the same author [18]. Using this criterion, GDM is identified if any of the following values are met or exceeded: fasting (5.1 mmol/L); 1st hour (10.0 mmol/L); and 2nd hour (8.5 mmol/L). Lipid profile parameters tested include total cholesterol (TC), triglycerides (TAG), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), and very low-density lipoprotein cholesterol (VLDL-C). A coupled enzymatic assay was used to determine the levels of TC and TAG in the serum. For HDL determination, a precipitation technique using polyethylene glycol in a glycine buffer was used. The concentration of LDL-C and VLDL-C were derived following Friedewald’s equation. Lastly, HbA1c levels in whole blood were tested using a boronate affinity assay.
Genomic DNA extraction, purification, and genotyping
Buffy coat from anticoagulated whole blood was isolated after spinning the samples at 2500 RPM for 10 minutes at room temperature. Genomic DNA was then extracted using the QIAamp DNA Blood Mini Kit with modifications. The quality of the extracted DNA was assessed using a combination of UV-Vis NanoDrop spectrophotometry and agarose gel electrophoresis. Qiagen Rotor-Gene Q5 Plex real-time PCR was used for gene amplification. The Pro12Ala polymorphism was genotyped using TaqMan genotyping assay. The context sequence used with the [VIC/FAM] dye to detect this was: AACTCTGGGAGATTCTCCTATTGAC[C/G]CAGAAAGCGATTCCTTCACTGATAC.
Ethical considerations
Ethical approval for this study is covered by the clearance given to the project entitled “Blood and Placental Gene Expression in Gestational Diabetes Mellitus: Potential Identification of Early Biomarkers” by the University of Santo Tomas Graduate School – Ethics Review Committee (E-2016-02-R3). Informed consent was obtained from the participants to ensure the voluntariness of participation. All participant identifiers were removed during the processing of samples and data analysis.
Statistical analysis
A combination of Microsoft Excel 2016, MedCalc free online calculator, and SPSS version 23 were used in the study. Mann-Whitney U-test was used to determine if there was a significant difference in various physical and biochemical characteristics between two study groups. Point-biserial correlation, on the other hand, was used to determine the association of the rs1801282 genotypes and alleles with the various physical and biochemical characteristics of the participants. Whereas, Pearson’s chi-square was used to determine the association of the said polymorphism with GDM development. Lastly, odds ratio computation was used to determine the likelihood of developing GDM depending on the pregnant women’s genotypic and allelic characteristics.
Results And Discussion
Physical and biochemical characteristics of GDM and non-GDM participants
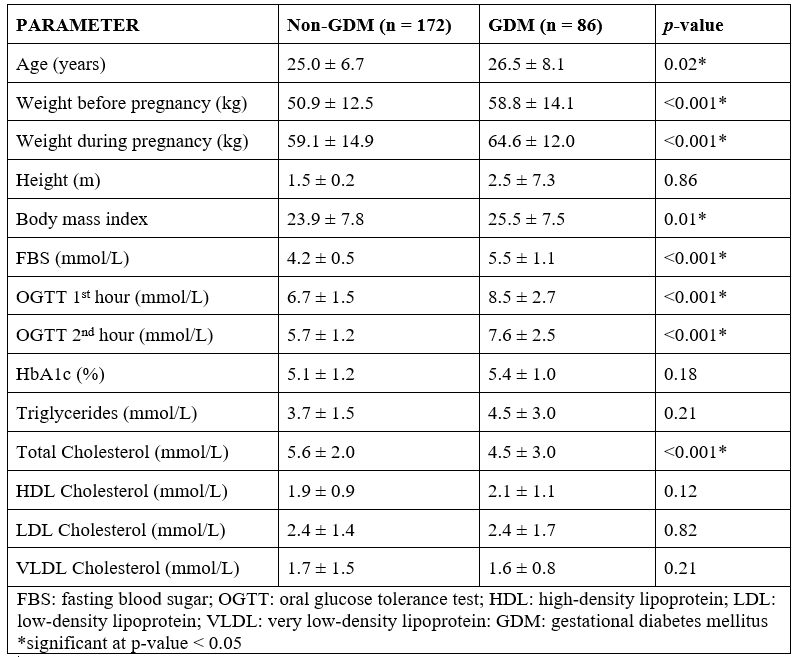
Table 1 illustrates the different parameters between non-GDM and GDM patients recruited. Variables such as age (p = 0.02), weight before pregnancy (p < 0.001), weight during pregnancy (p < 0.001), BMI (p < 0.001), fasting blood sugar (FBS) (p < 0.001), OGTT 1st hour (p < 0.001), and OGTT 2nd hour (p < 0.001) were significantly higher among the GDM group, which may indicate that factors such as age and BMI are strong predictors of GDM development. On the other hand, TC levels (p < 0.001) were significantly higher among the non-GDM group compared to the GDM group. These findings were consistent with findings from previous studies. Based on the study of Makgoba et al. (2011), similar results showed that development of GDM has a strong positive association with advancing maternal age [19]. Another study indicated that the risk of GDM becomes significantly and progressively increased from 25 years onwards [20]. With regards to maternal BMI, Kim et al. in 2010 attributed a large percentage of GDM cases were related to having overweight and obese BMI [21, 22]. Moreover, the study conducted by Cavicchia et al. in 2014 suggests an increased risk of GDM development among overweight, obese, and extremely obese women [23]. Further studies showed that excessive weight gain is considered as a risk factor for GDM development [24], and women with GDM have a higher BMI compared to those without [25].
Table 1: Mann-Whitney U-test analysis of the physical and biochemical characteristics of non-GDM and GDM participants.
Association of the rs1801282 SNP with various GDM-associated parameters
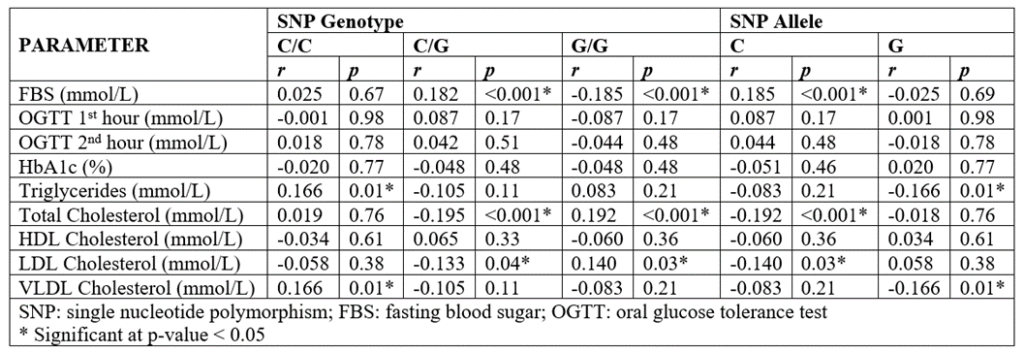
As shown in Table 2, the C/G genotype, G/G, and the C allele are significantly correlated with FBS, TC, and LDL-C levels. Both the C/G genotype and the C allele show a positive correlation with the FBS levels, therefore suggesting that its presence increases FBS levels. For the TC and LDL-C levels, the C/G genotype and the C allele show a negative correlation, therefore, suggesting that its presence decreases the abovementioned parameters. The G/G genotype showed a positive correlation with both TC and LDL-C levels and a negative correlation with FBS levels. Similar results were seen in a previous study conducted by Bego et al. (2011) wherein, an increased level of TC and LDL-C levels are observed among those with G/G genotype [26]. Also, other studies suggest that the SNP causes an impaired clearance of LDL-C and VLDL-C that results in elevated serum levels of TC, LDL-C, and VLDL-C [27] which is consistent with the present study. However, in some studies, the C/C and C/G genotypes of the rs1801282 SNP has been found to have a protective role [28, 29] against GDM development which is contradictory with our findings.
Table 2 also suggests that the C/C genotype and the G allele are both significantly correlated with TAG and VLDL-C levels. The C/C genotype showed a positive correlation suggesting that its presence significantly increases the said lipoprotein levels. The G allele, on the other hand, showed a negative correlation with both TAG and VLDL-C levels indicating that its presence significantly decreases the levels of the two. These results were consistent with the study conducted by Muñoz-Yañez et al. in 2016, AlSaleh et al. in 2011 and Huang et al. in 2011, wherein they observed a negative correlation between the Pro12Ala polymorphism and triglyceride levels [30–32]. No single mechanism can fully explain this association. In the study of Hugeunin et al. (2010), the Pro12Ala polymorphism is associated with decreased transcriptional activity and increased insulin sensitivity among Caucasians. The increased sensitivity to insulin promotes fat deposition in adipose tissue and to the decreased levels of TAG in serum [33]. Another suggested mechanism is the role of PPARG in the metabolic pathway. CD36 is a receptor for long-chain fatty acids and is a direct up-regulation target gene of PPARG. CD36 function as a transporter of fatty acids in adipose tissue, which facilitates fatty acid uptake and lipid accumulation in adipocytes [34].
Table 2: Point-biserial correlation of the rs1801282 genotype and allele with various GDM-associated parameters.
Association of the rs1801282 SNP with GDM
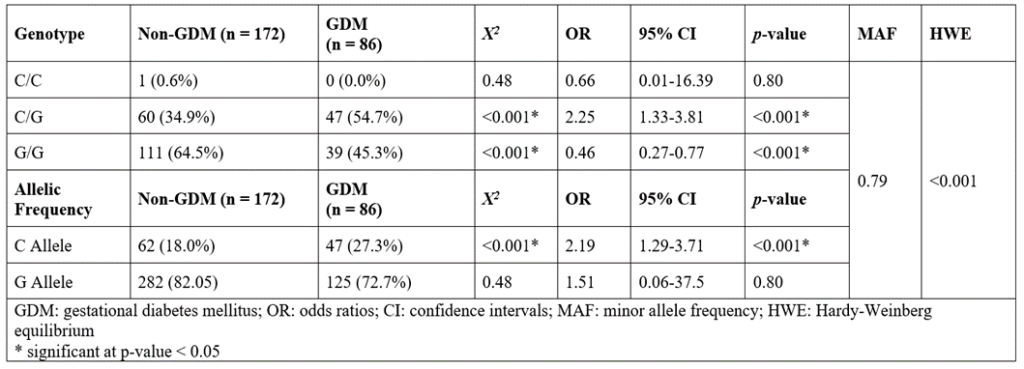
Table 3 summarizes the result for the association of the SNP with GDM. Also, the table shows the minor allele frequency (0.79) and the results of the Hardy-Weinberg equilibrium computation (p < 0.001). As shown in Table 3, the majority of non-GDM pregnant women have a G/G genotype (64.5%) as compared to the GDM group (45.4%). On the other hand, the majority of pregnant women with GDM have a C/G genotype (54.7%). As for the alleles, the G allele was observed in both groups (99.4%-100%). These results suggest that the presence of G allele and the G/G genotype is associated with a reduced incidence of GDM among the participants. As for the likelihood of GDM development, results are also summarized in Table 3. The individual odds ratios suggest that that the presence of the heterozygous C/G genotype (OR:2.25; 95% CI: 1.33-3.81; p < 0.001) and the ancestral C allele (OR:2.19; 95% CI: 1.29-3.71; p < 0.001) significantly increases the likelihood of developing GDM. On the other hand, the presence of G/G genotype (OR:0.46; 95% CI: 0.27-0.77; p < 0.001) showed a significant protective role against GDM development.
Table 3: Pearson’s chi-square analysis and odds ratio computation analysis of the association of the rs1801282 genotypes and allelic frequencies with GDM.
Results of the present study were similar from the studies of Stuebe et al. (2014) and Kawai et al. (2017), where an increased risk of developing GDM was observed among pregnant women with the C allele [35, 36]. In a study conducted by Zhang et al. in 2013, they indicated that the C allele of the rs7754840 SNP was significantly associated with risk of GDM while the G allele of rs1801282 SNP was not significantly associated with the possibility of developing GDM [37]. Furthermore, in a study by Zhu et al. in 2017, they were able to show that the polymorphism in the gene increases the risk of T2DM development [38]. Lastly, a study conducted by Lin et al. in 2018 pointed out that after pooling data from eight studies that included Asian pregnant women, the mutant G allele of the rs1801282 polymorphism was significantly associated with decreased GDM risk under the dominant model (GG + GC vs CC), heterozygous model (GC vs. CC), and allele model (G allele vs. C allele), but not under the recessive and homozygous models [39]. A possible reason for these result is the interaction between the PPARG genotype and circulating lipids on insulin resistance (IR) as proposed by Stryjecki et al. [10]. Lower fasting insulin concentration and insulin sensitivity are observed among carriers of the Ala12 allele when TC levels are high. Carriers of this allele were found to be insulin sensitive despite high levels of circulating LDL-C, further suggesting the protective role of the polymorphism against IR development despite the presence of dyslipidemia [40]. Moreover, another possible reason is that the PPARG also regulates genes involved in insulin signaling and the expression of proinflammatory cytokines [41,42].
The biochemical profile between pregnant women with the G/G genotype versus those with the C/C and C/G genotypes
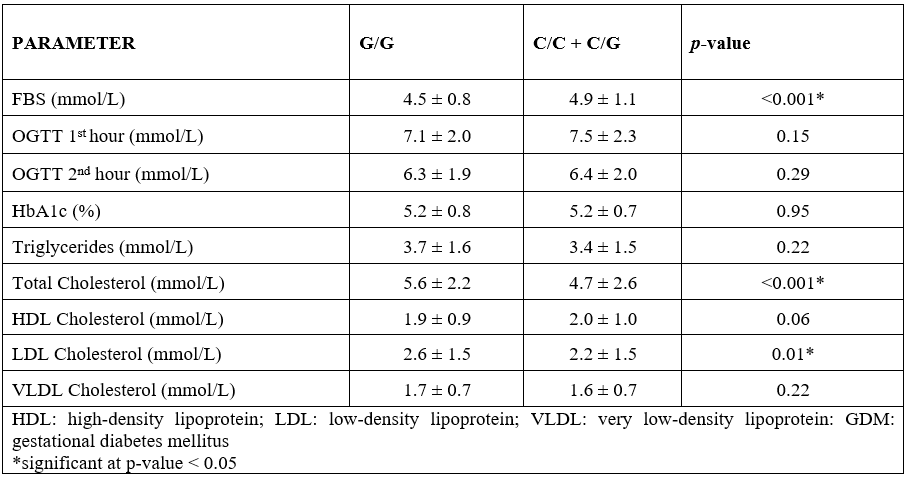
In order to determine the influence of the SNP with the biochemical profile of the pregnant women, participants were sub-grouped based on their genotypes (G/G vs. C/C + C/G). The results are summarized in Table 4. Based on the results, FBS is significantly higher (p < 0.001) among those with the C/C + C/G genotypes compared to those with the G/G genotype. On the other hand, TC (p < 0.001) and LDL-C (p = 0.01) levels are higher among those with the G/G genotype. These results are in agreement with a study conducted by Franzago et al. in 2017, where they found out that the rs1801282 SNP correlates with LDL-C levels among pregnant women on their third trimester [43]. Other studies by Plescovic et al. (2016) and Alves et al. (2017) indicated that the mutant allele of the rs1801282 SNP is associated with high levels of TAG and LDL-C [8, 44]. As explained by Jiang et al. in 2017, the high levels of TAG and LDL-C brought by the SNP leads to impaired cholesterol homeostasis, obesity, and especially the development of diabetes mellitus [45]. The relationship between the rs1801282 SNP with lipid profile levels may be explained by the importance of the PPARG in the regulation of genes encoding for proteins involved in adipocyte differentiation and lipid storage [46].
Table 4: Mann-Whitney U-test analysis of the lipid profile levels between pregnant women with the G/G genotype versus those with C/C and C/G genotypes.
Conclusion
To our knowledge, this is the first study that determined the association of the rs1801282 SNP (Pro12Ala or CCA to GCA) in the PPARG gene with GDM among Filipinos. Previous studies explicitly indicated the genetic variability depending on ethnicity of polymorphism studies. Based on our results, among the pregnant Filipino women enrolled in this study, the presence of the C allele and the heterozygous C/G genotype were found to be significantly associated with an increased likelihood of GDM development. Meanwhile, the presence of the G/G genotype showed a significant protective role against GDM. Furthermore, the presence of the G/G genotype corresponds to lower levels of FBS but higher levels of TC and LDL-C. On the other hand, the C/C genotype is related to higher levels of TAG and VLDL-C while the C/G genotype is correlated with higher levels of FBS and lower levels of TC and LDL-C.
Declarations
Funding; This research is partially funded by the Department of Science and Technology – Philippine Council for Health Research and Development under the project entitled “Blood and Placental Gene Expression in Gestational Diabetes Mellitus: Potential Identification of Early Biomarkers.”
Conflicts of interest: Authors declare no conflict of interest.
Ethics approval: Study has an approved ethical clearance under the project – Blood and placental gene expression in gestational diabetes mellitus: potential identification of early biomarkers (E-2016-02-R3).
Consent to participate: Participants of this study were asked to sign informed consent forms voluntarily.
Consent for publication: All data presented here are primary data gathered in the study.
Availability of data and material. Not applicable
Code availability: Not applicable
References
- Mitanchez D, Burguet A, Simeoni U (2014) Infants born to mothers with gestational diabetes mellitus: Mild neonatal effects, a long-term threat to global health. J Pediatr 164:445–450.
- Ma RCW, Tutino GE, Lillycrop KA, et al (2015) Maternal diabetes, gestational diabetes and the role of epigenetics in their long term effects on offspring. Prog Biophys Mol Biol 118:55–68.
- Kampmann U (2015) Gestational diabetes: A clinical update. World J Diabetes 6:1065.
- Li Q, Chen R, Bie L, et al (2015) Association of the variants in the PPARG gene and serum lipid levels: A meta-analysis of 74 studies. J Cell Mol Med 19:198–209.
- Ryckman KK, Spracklen CN, Smith CJ, et al (2015) Maternal lipid levels during pregnancy and gestational diabetes: A systematic review and meta-analysis. BJOG An Int. J. Obstet. Gynaecol. 122:643–651.
- Kruzliak P, Haley AP, Starcevic JN, et al (2015) Polymorphisms of the Peroxisome Proliferator-Activated Receptor-γ (rs1801282) and its coactivator-1 (rs8192673) are associated with obesity indexes in subjects with type 2 diabetes mellitus. Cardiovasc Diabetol 14.
- Hasan NS, Kamel SA, Hamed M, et al (2017) Peroxisome proliferator-activated receptor-γ polymorphism (rs1801282) is associated with obesity in Egyptian patients with coronary artery disease and type 2 diabetes mellitus. J Genet Eng Biotechnol 15:409–414.
- Alves MC, de Morais CC, Augusto EM, et al (2017) Polymorphisms in PPARG and APOE: relationships with lipid profile of adolescents with cardiovascular risk factors. Nutrire 42.
- Chan KHK, Niu T, Ma Y, et al (2013) Common genetic variants in peroxisome proliferator-activated receptor-γ (PPARG) and type 2 diabetes risk among women’s health initiative postmenopausal women. J Clin Endocrinol Metab 98.
- Stryjecki C, Peralta-Romero J, Alyass A, et al (2016) Association between PPAR-γ 32 Pro12Ala genotype and insulin resistance is modified by circulating lipids in Mexican children. Sci Rep 6.
- Johansson LE, Danielsson P, Norgren S, et al (2009) Interaction between PPARG Pro12Ala and ADIPOQ G276T concerning cholesterol levels in childhood obesity. Int J Pediatr Obes 4:119–125.
- Hsiao TJ, Lin E (2015) The Pro12Ala polymorphism in the peroxisome proliferator-activated receptor gamma (PPARG) gene in relation to obesity and metabolic phenotypes in a Taiwanese population. Endocrine 48:786–793.
- Shanmuga Priya S, Sankaran R, Ramalingam S, et al (2016) Genotype phenotype correlation of genetic polymorphism of PPAR gamma gene and therapeutic response to pioglitazone in type 2 diabetes mellitus- a pilot study. J Clin Diagnostic Res 10:FC11–FC14.
- Al-Naemi AH, Ahmad AJ (2018) Is the rs1801282 (G/C) polymorphism of PPAR – gamma gene associated with T2DM in Iraqi people? Open Access Maced J Med Sci 6:447–455.
- Trombetta M, Bonetti S, Boselli ML, et al (2013) PPARG2 Pro12Ala and ADAMTS9 rs4607103 as “insulin resistance loci” and “insulin secretion loci” in Italian individuals. the GENFIEV study and the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS) 4. Acta Diabetol 50:401–408.
- Tellechea ML, Aranguren F, Pérez MS, et al (2009) Pro12Ala Polymorphism of the Peroxisome ProliferatorActivated Receptor-γ Gene is Associated With Metabolic Syndrome and Surrogate Measures of Insulin Resistance in Healthy Men. Circ J 73:2118–2124.
- Watanabe RM, Black MH, Xiang AH, et al (2007) Genetics of Gestational Diabetes Mellitus and Type 2 Diabetes. Diabetes Care 30:S134–S140.
- Pineda-Cortel MRB, M Manalo ME, C Canivel RR, et al (2018) Screening and Diagnosis of Gestational Diabetes Mellitus Using 75-g Oral Glucose Tolerance Test Following the WHO, ADA, and IADPSG Criteria. J Diabetes Metab 09:7–10.
- Makgoba M, Savvidou MD, Steer PJ (2012) An analysis of the interrelationship between maternal age, body mass index and racial origin in the development of gestational diabetes mellitus. BJOG An Int J Obstet Gynaecol 119:276–282.
- Lao TT, Ho L-F, Chan BCP, Leung W-C (2006) Maternal Age and Prevalence of Gestational Diabetes Mellitus. Diabetes Care 29:948–949.
- Kim SY, England L, Wilson HG, et al (2010) Percentage of gestational diabetes mellitus attributable to overweight and obesity. Am J Public Health 100:1047–1052.
- Pawlik A, Teler J, Maciejewska A, et al (2017) Adiponectin and leptin gene polymorphisms in women with gestational diabetes mellitus. J Assist Reprod Genet 34:511–516.
- Cavicchia PP, Liu J, Adams SA, et al (2014) Proportion of gestational diabetes mellitus attributable to overweight and obesity among non-hispanic black, non-hispanic white, and hispanic women in South Carolina. Matern Child Health J 18:1919–1926.
- Petrova Genova M, Dimitrova Atanasova B, Nikolova Todorova-Ananieva K (2019) Body Mass Index and Insulin Sensitivity/Resistance: Cross Talks in Gestational Diabetes, Normal Pregnancy and Beyond. In: Body-mass Index and Health. IntechOpen.
- Layton J, Powe C, Allard C, et al (2019) Maternal lipid profile differs by gestational diabetes physiologic subtype. Metabolism 91:39–42.
- Bego T, Dujic T, Mlinar B, et al (2011) Analysis of PPARG gene polymorphisms in patients with metabolic syndrome and type 2 diabetes from Bosnia and Herzegovina. FEBS J 278:175.
- Nasereddin A, Ereqat S, Azmi K, et al (2009) Impact of the pro12Ala polymorphism of the PPAR-gamma 2 gene on metabolic and clinical characteristics in the palestinian type 2 diabetic patients. PPAR Res.
- Morini E, Tassi V, Capponi D, et al (2008) Interaction between PPARγ2 variants and gender on the modulation of body weight. Obesity 16:1467–1470.
- Bhatt SP, Misra A, Sharma M, et al (2012) Ala / Ala Genotype of Pro12Ala Polymorphism in the Peroxisome Proliferator-Activated Receptor – γ2 Gene Is Associated with Obesity and Insulin Resistance in Asian Indians. Diabetes Technol Ther 14:828–834.
- Muñoz-Yáñez C, Pérez-Morales R, Moreno-Macías H, et al (2016) Polymorphisms FTO rs9939609, PPARG rs1801282 and ADIPOQ rs4632532 and rs182052 but not lifestyle are associated with obesity related-traits in Mexican children. Genet Mol Biol 39:547–553.
- AlSaleh A, O’Dell SD, Frost GS, et al (2011) Interaction of PPARG Pro12Ala with dietary fat influences plasma lipids in subjects at cardiometabolic risk. J Lipid Res 52:2298–2303.
- Huang X, Zhao J, Zhao T (2011) Effects of peroxisome proliferator activated receptor-gamma 2 gene Pro12Ala polymorphism on fasting blood lipids: A meta-analysis. Atherosclerosis 215:136–144.
- Huguenin GVB, Rosa G (2010) The Ala allele in the PPAR-γ2 gene is associated with reduced risk of type 2 diabetes mellitus in Caucasians and improved insulin sensitivity in overweight subjects. Br J Nutr 104:488–497.
- Meirhaeghe A, Fajas L, Helbecque N, et al (2000) Impact of the Peroxisome Proliferator Activated Receptor γ2 Pro12Ala polymorphism on adiposity, lipids and non-insulin-dependent diabetes mellitus. Int J Obes 24:195–199.
- Kawai VK, Levinson RT, Adefurin A, et al (2017) A genetic risk score that includes common type 2 diabetes risk variants is associated with gestational diabetes. Clin Endocrinol (Oxf) 87:149–155.
- Stuebe AM, Wise A, Nguyen T, et al (2014) Maternal genotype and gestational diabetes. Am J Perinatol 31:69–76.
- Zhang C, Bao W, Rong Y, et al (2013) Genetic variants and the risk of gestational diabetes mellitus: A systematic review. Hum Reprod Update 19:376–390.
- Zhu L, Huang Q, Xie Z, et al (2017) PPARGC1A rs3736265 G>A polymorphism is associated with decreased risk of type 2 diabetes mellitus and fasting plasma glucose level. Oncotarget 8.
- Lin PC, Chou PL, Wung SF (2018) Geographic diversity in genotype frequencies and meta-analysis of the association between rs1801282 polymorphisms and gestational diabetes mellitus. Diabetes Res. Clin. Pract. 143:15–23.
- Kubota N, Terauchi Y, Miki H, et al (1999) PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell 4:597–609.
- Usuda D (2014) Peroxisome proliferator-activated receptors for hypertension. World J Cardiol 6:744.
- Bensinger SJ, Tontonoz P (2008) Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 454:470–477.
- Franzago M, Fraticelli F, Nicolucci A, et al (2017) Molecular Analysis of a Genetic Variants Panel Related to Nutrients and Metabolism: Association with Susceptibility to Gestational Diabetes and Cardiometabolic Risk in Affected Women. J Diabetes Res 2017:1–7.
- Pleskovič A, Šantl Letonja M, Cokan Vujkovac A, et al (2016) Polymorphisms of the PPAR- γ (rs1801282) and Its Coactivator (rs8192673) Have a Minor Effect on Markers of Carotid Atherosclerosis in Patients with Type 2 Diabetes Mellitus. PPAR Res 2016:.
- Jiang J, Xie Z, Guo J, et al (2017) Association of PPARG rs 1801282 C>G polymorphism with risk of colorectal cancer: from a case-control study to a meta-analysis. Oncotarget 8:
- Lee WS, Kim J (2015) Peroxisome proliferator-activated receptors and the heart: Lessons from the past and future directions. PPAR Res. 2015.


