Characterization of the Subtypes of HLA-B*35 Alleles Detected in Patients with Mild and Severe Infection with Andes Hantavirus in Chile Pablo A. Ferrer Campos*
Laboratorio de Medicina Molecular, Hospital Clínico Universidad de Chile, Santiago, Chile
*Correspondence to: Pablo A. Ferrer Campos
Citation: Ferrer P (2022) Characterization of the Subtypes of HLA-B*35 Alleles Detected in Patients with Mild and Severe Infection with Andes Hantavirus in Chile. Sci Academique 3(1): 19-25
Received: 02 April, 2021; Accepted: 02 June 2022; Publication: 10 June 2022
Abstract
Background: In Chile the infection caused by the Andes Hantavirus (ANDV) has a variable clinical expression ranging from asymptomatic, mild to severe cases.
Objective: The objective of this work was the characterization of the subtypes HLA-B*35 alleles using high resolution SSP-PCR.
Method: Molecular determination of subtypes HLA-B*35 alleles was realized by high resolution SSP-PCR utilizing the B*35 SSP UniTray® Kit (Invitrogen).
Results: We determinate twenty three out of twenty four (23/24, 95.8%) possible subtypes HLA-B*35 alleles detected in 87 patients infected with ANDV. One subtype HLA-B*35 allele (1/24, 4.2%) could not be characterized for this technology and it was classified as HLA-B*35??. The HLA-B*3505 and HLA-B*3508 alleles were found exclusively in the patients with mild clinical course of ANDV infection, while the HLA-B*35?? allele, was detected only in the group of patients with severe symptoms. The HLA-B*3501, HLA-B*3502, HLA-B*3509, HLA-B*3520 y HLA-B*3543 alleles were found in both groups of patients.
Conclusion: High resolution SSP-PCR allowed the characterization of seven subtypes of HLA-B*35 alleles. However, with the kit utilized was not possible resolve the HLA-B*35?? allele. In this preliminary study we can observe that subtypes HLA-B*35 alleles are stratified according with clinical course of ANDV infection in Chile. This is very valuable information since it could be used to open a field of research in the development of vaccines against Andes Hantavirus. For characterize the HLA-B*35?? allele we propose using sequence-based typing (SBT) due to that probably the sequences of the exons two and three of this allele are not included in the commercial kit utilized in this work.
Background
Hantaviruses, were first recognized during the Korean War in the early 1950 as the etiologic agent of Korean hemorrhagic fever, reclassified as hemorrhagic fever with renal syndrome (HFRS) [1]. In 1993, a newly recognized specie of hantavirus was found to be the causing agent of the Hantavirus Cardiopulmonary Syndrome (HCPS) caused by the Sin Nombre Virus (SNV) in New Mexico and other Four Corners states in the United States of America [2]. In addition, to Hantaan virus and SNV, several other hantaviruses have been implicated as the etiologic agents for either HFRS or HCPS [3]. In Chile, HCPS caused by the Andes virus (ANDV) infection, has a mortality rate of 34% [4,5]. In ANDV infection we can observe asymptomatic, mild, and severe cases [6]. The severe cases are characterized by the development of respiratory insufficiency, circulatory failure and cardiogenic shock similar to HCPS due to SNV [7]. Although the pathogenesis of HFRS or HCPS is little known, there is evidence to suggest that the disease is caused by damage of the host immune system, rather than the direct lytic effect of the virus [8]. Immunopathology is critical in the hantaviral diseases, therefore we expect that the HLA system may be playing a key role [9]. To this respect previously has been found that HLA-B8-DR3 haplotype is associated with severe outcome of Puumala virus (PUUV) infection [10]. Afterwards, the association of fatal HCPS or HCPS complicated by shock with HLA-B*35 allele was reported [11]. More recently, it has been reported that HLAB*3501 allele is associated with severe HCPS caused by SNV infection implying the involvement of CD8+ cytotoxic T cells [12].
In this direction we carried out a research to know if the HLA alleles are stratified with the clinical course of ANDV infection. For this purpose, we studied 87 individuals infected with ANDV separated in patients with mild and severe clinical course. We found 24 individuals carrying the HLA-B*35 alleles (27.6%) and this allele was more frequent in benign than severe cases of infection [13]. The prevalence of HLA-B*35 alleles was similar to the prevalence of 26-27% in general population of Chile [14].
Due to ANDV is not a lytic virus is possible to hypothesize in favor of a pathogenetic role of the host’s immune response in the ANDV infection as the result of an excessive reaction immune system against virus [8,9,12,15]. The human HLA system is implicated in two crucial biological events: the immune response to pathogens and the graft/transplant rejection [16-20]. Therefore, the focus of our interest is to understand the role of the human HLA system in the resulting clinical course following the infection with ANDV [10,13,21].
Objectives
In the present work we addressed the study of the differences at molecular level of the HLA-B*35 alleles detected in patients with mild and severe infection by ANDV.
Study Design
Were studied 24 individuals with ANDV infection confirmed by the detection of antibodies against ANDV [13,22,23]. Human genomic DNA was isolated from peripheral blood using the Lahiri’s method [24]. Patients were genotyped for determinate the subtypes HLA-B*35 alleles using SSP PCR B*35 SSP UniTray® Kit (Invitrogen) according to manufacturer’s instructions [16].
Results
We achieved characterize twenty three out of twenty-four (95.8%) patients carrying the HLA-B*35 allele [13]. One HLA-B*35 allele (4.2%) could not be characterized and was designed as HLA-B*35?? allele. We found eight different subtypes of HLA-B*35 alleles including the uncharacterized allele. Interestingly, HLA-B*3505 and HLA-B*3508 alleles were found exclusively in the mild patients group. HLA-B*3501, HLA-B*3502, HLA-B*3509, HLA-B*3520 y HLA-B*3543 alleles were found in both groups (Figure 1). Finally, uncharacterized HLA-B*35?? allele was detected only in the severe patients group. Uncharacterized HLA-B*35?? allele correspond to a limitation of SSP-PCR technology and is probably it is due to high polymorphism of HLA molecules. In this sense, one probability is that sequences of this allele were not included in the high resolution SSP PCR kit used. Alternatively, uncharacterized HLA-B*35?? allele might be a new HLA-B*35 subtype. Unfortunately, we sequenced the HLA-B*35?? allele for HLA typing by sequence-based typing (SBT) of exons II and exon III of HLA-B molecules but the sequences obtained were not of good quality and we were unable to genotype this allele [25-27,33].
Discussion
Hantavirus disease in Chile is considered a rare disease due to the low number of cases per year.
Therefore, despite this limitation our results are highly suggestive because they indicate that there is a stratification of subtypes of HLA-B *35 alleles with the course of infection by ANDV. This preliminary study also allows to explain the discrepancies between the associations of HLA-B * 3501 allele with the clinical course of HCPS caused by ANDV and SNV [12,13,15,29].
With respect to HLA-B*35 alleles and their association with infection diseases, we focus on B*35 alleles because other researchers have published important works on the B*35 allele and association with HIV and Hantavirus. For example, HIV has been showed that HLA-B*35 allele was associated with more rapid progression to AIDS, greater infectivity, and higher viral load [10,13,25, 28,29,35].
With respect to HCPS caused by ANDV, we detected HLA-B*35 alleles were found more frequently in patients with mild disease than in patients with severe disease [13] (Figure 1). On the other hand, in USA contrary to the study carried out for us, the HLA-B*3501 allele was associated with increased risk for developing severe HCPS caused by SNV (12, 15). For this reason and in an attempt for resolve this discrepancy we realized the characterization of HLA-B*35 subtypes detected in our study and this way know which HLA-B*35 subtypes alleles are stratified with clinical course of ANDV infection (Figure 1). To this respect in the Figure 2 we can observe evident structural differences in the peptide-binding groove of HLA-B*35 alleles. The differences observed corresponding to changes in the amino acids in the zone of the HLA molecule where the subtypes of HLA-B*35 alleles are defined. In fact, the HLA-B*3508 allele, has an Arg 156, compared to Leu 156 in the HLA-B*3501 allele. On the other hand, HLA-B*3505 allele differ by three amino acids: Thr 94, Leu 95, and Ser 97 compared to HLA-B*3501 molecule, with Ile 94, Ile 95 and Arg 97. All these chemicals differences in the amino acid sequences must lead to profound functional changes in antigen-binding activity and CTL function and possibly with the clinical course of the infectious disease. Since the differences of HLA-B*3505, HLA-B*3508 with respect to HLA-B*3501 are related to the primary anchor residue binding pockets we can speculate that such primary structure variability can have a major impact on determinant selection in antiviral T cell responses against ANDV [34,36,37].
Figure 1: In this figure we show the subtypes of HLA-B*35 alleles detected in this study. The A group (n=14) corresponding to subtypes detected in the mild patients group and the B group (n=10) corresponding to the subtypes detected in the severe patients group. The intersection area corresponding to the alleles in common to both groups. We can see that HLA-B*3505 and HLA-B*3508 alleles were detected only in the mild patients group (A group) while one uncharacterized HLA-B*35?? allele was detected only in the severe patients group (B group).
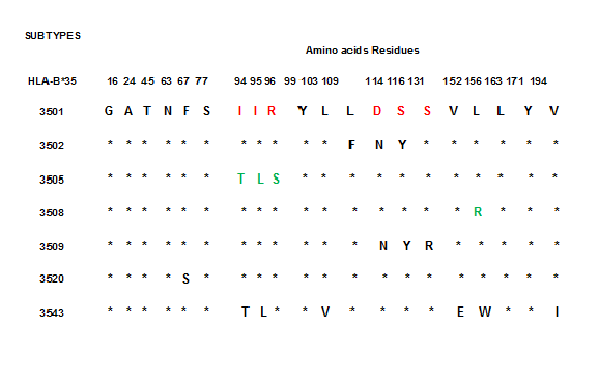
Figure 2. This figure shows different subtypes of HLA-B*35 alleles. The HLA-B*3505 is defined for the residues T, L and S in the positions 94, 95, and 96. The HLA-B*3508 is defined for the R residue in the position 156 and the HLA-B*3509 allele is defined for the residues N, Y and R in the positions 114, 116, and 131 in relation to HLA-B*3501 allele in the same positions. These subtypes of HLA-B*35 allele were found in patients infected with Andes Hantavirus in Chile. We can observe that the changes in these HLA-B*35 alleles are limited to nucleotides encoding for amino acid in the peptide-binding groove or in residues related to this function in the HLA protein. Therefore, the sequences changes in the subtypes of HLA-B*35 allele lead to profound changes in antigen-binding activity and consequently may be affect the clinical course of ANDV infection.
These human HLA alleles present different amino acids inside or outside of the peptide-binding groove, therefore, it is possible that they may have different peptide binding profiles and consequently eliciting different immune response to ANDV [38].
In this sense result suggestive identify which are the epitopes of ANDV selected for the HLA-B*3505 and HLA-B*3508 alleles and probe these peptides as a potentially vaccine for ANDV infection [39].
However, despite these suggestive results on the participation of host genetics in hantavirus infection, we think there non genetically associated variables possibly involved in the individual susceptibility to ANDV infection we are inclined to propose early diagnosis, time and conditions of patients’ transfers, quality of medical support, strain virulence, viral load, entry mechanism and inhaled inoculum’s size of hantavirus [13].
Additionally, for hantavirus disease is unknown how much of each of these factors may contribute in facilitating or impeding the hantavirus infection in humans in Chile and America [22,23,30-32] Finally, other genetic factors, in the form of genes or genetic regulatory elements, part of the human genome that may be involved in conferring susceptibility or resistance to this particular virus.
Although ours study included only a small number of patients, the preliminary results presented in this report, allows us to infer that exist an important participation of immunogenetic phenomenon can occur in ANDV infection and the reported findings certainly grant the need to perform a study with a larger population sample [13,15].
The factors controlling epitope selection in the T cell response to viruses are not fully understood. In this investigation we have found two subtypes of HLA-B*35 alleles: HLA-B*3505 and HLA-B*3508 alleles that may be binding peptides derived from ANDV. Nevertheless, we believe that our study represents a genuine effort to shed some light on the intricate medical and scientific enigma involving the ANDV infection and its relationship with its host immune system, including the HLA system. Future studies of the human HLA system variability and ANDV infection of its human host should certainly include a larger cohort of patients and perhaps an improved algorithm of clinical criteria to appropriately grading the severity of infection. Moreover, we should probably apply stricter anthropological criteria to correctly identify the ethnical background of each patient included in the patient population sample. Lastly, we need to realize studies of peptide-HLA association with an appropriate peptide pool derived from ANDV and molecular modeling to confirm our theory regarding the structural effects of amino acids changes in the HLA molecules and how such structural variations may affect the clinical evolution of ANDV infection in the human host. Altogether, these studies will be instrumental in designing a more rational approach for the development of more effective drugs and vaccines against ANDV [39].
Disclosure
Funding: This study was supported by Scholarship to Pablo Ferrer Campos.
Competing interest: No conflict of interest.
Ethical Approval: Each subject received information about the study and written informed consent was obtained. This protocol was approved for the ethical committee of Faculty of Medicine Universidad de Chile.
Acknowledgement: Proyecto Hantavirus Ecología y Enfermedad.
References
- Lee HW, Lee PW, Johnson KM (1978) Isolation of the etiologic agent of Korean Hemorrhagic fever. J Infect Dis 137: 298-308.
- Centers for Disease Control and Prevention (CDC) (1993) Outbreak of acute illness-southwestern United States, 1993. MMWR Morb Mortal Wkly Rep 42: 421-424.
- Muranyi W, Bahr U, Zeier M, Van der Woude FJ (2005) Hantavirus Infection. J Am Soc Nephrol 16: 3669-3679.
- Sotomayor V (2007) Vigilancia de enfermedad por hantavirus. El Vigía 10:24-27.
- Pinna DM, Martinez VP, Bellomo CM, López C, Padula P (2004) Nueva Evidencia epidemilógica y molecular a favor de la transmisión interhumana para el linaje sout del hantavirus Andes. Medicina Buenos Aires 64: 43-46.
- Riquelme R, Riquelme M, Torres A, Rioseco ML, Vergara JA, et al. (2003) Hantavirus Pulmonary Syndrome, Southern Chile. Emerg Infect Dis 9: 1438-1443.
- Toro J, Vega J, Khan A, Mills J, Padula P, et al (1998) An Outbreak of hantavirus pulmonary syndrome, Chile, 1997. Emerg Infect Dis 4: 687-694.
- Terajima M, Hayasaka D, Maeda K, Ennis F (2007) Immunopathogenesis of hantavirus pulmonary syndrome and hemorrhagic fever with renal syndrome: Do CD8+T cells trigger capillary leakage in viral hemorrhagic fevers?. Immunol Lett 15: 117-120.
- Easterbrook JD, Klein SL (2008) Immunological mechanisms mediating hantavirus persistence in rodent reservoirs. Plos Pathogens 4: e10001721.
- Mustonen J, Partanen J, Kanerva M, Pietila K, Vapalahti O, et al. (1996) Genetic susceptibility to severe course of nephropathia epidemica caused by puumala hantavirus. Kidney Int 49: 217-221.
- Mertz G, Hjelle B, Williams T, Koster F, (1998) Emergence and control of rodent-borne viral diseases (hantaviral and arenal diseases). Emerging Disease. In: Saluzzo JF, Dodet B, editors. Chapter: Host responses in the hantavirus cardiopulmonary syndrome. Annecy: Foundation Marcel Merieux. pp. 131-137.
- Kilpatrick E, Terajima M, Koster F, Catalina M, Cruz J, et al. (2004) Role of Specific CD8+ T cells in the severity of a fulmianant zoonotic viral hemorrhagic fever, Hantavirus Pulmonary Syndrome. J Immunol 172: 3297-3304.
- Ferrer P, Vial P, Ferrés M, Godoy P, Cuiza A, et al. (2007) Susceptibilidad genética a hantavirus Andes: Asociación entre la expresión clínica de la infección y alelos del sistema HLA en pacientes chilenos. Rev Chil Infect 24: 351-359http://www.scielo.cl/pdf/rci/v24n5/art01.pdf. Accessed 2009 Jun 10.
- Droguett MA, Oyarzún MJ, Alruiz P, Jerez V, Mezzano S, et al. (2005) Human leukocyte antigens in indigenous (Mapuche) people in a regional renal transplantation program in Chile. Transplantation Proceedings 37: 3367-3371.
- Koster F, Vigil J, Olson D, Terajima M, Ennis F, et al. (2001) Class I, II and III HLA alleles associated with severe hantavirus cardiopulmonary syndrome in the southwest US (meeting abstract). Am J Trop Med 65: 152.
- Powis SH, Vaughan RW (2000) MHC protocols. London: Humana Press.
- Klein J, Sato A (2000) The HLA system-First of two parts. N Engl J Med 343: 702-709.
- Klein J, Sato A (200) The HLA system-Second of two parts. N Engl J Med 343: 782-786.
- Shiina T, Inoko H, Kulski JK (2004) An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens 64: 631-649.
- Marsh SGE, Parham P, Barber LD (2000) The HLA FactsBook. London: Academic Press.
- Yin Ma, Jiuping Wang, Bin Yuan, Meiliang Wang, Yun Zhang, et al. (2013) HLA-A2 and B35 restricted Hantaan virus nucleoprotein CD8+ T-cell epitope-specific immune response correlates with milder disease in hemorrhagic fever with renal syndrome. PLOS Neglected Tropical Diseases 7: e2076.
- Castillo C, Sanhueza L, Tager M, Muñoz S, Ossa G et al. (2002) Seroprevalence of antibodies against hantavirus in 10 communities of the IX region of Chile where hantavirus infection were diagnosed. Rev Med Chile 130: 251-258.
- Tager M, Vial P, Castillo C, Godoy P, Hjelle B, et al. (2003) Hantavirus Prevalence in the IX region of Chile. Emerg infect Dis 9: 827-832.
- Lahiri DK, Nurnberger JI, (1991) A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nuncl Acids Res 19: 5444.
- Guo Z, Gatterman M, Hodd L, Hansen J, Petersdorf E (2001) Oligonucleotide arrays for high-throughput SNPs detection in the MHC Class I genes: HLA-B as a model system. Genome Res 12: 447-457.
- Helmberg W, Dunivin R, Feolo M (2004) The sequencing-based typing tool of dbMHC: typing highly polymorphic gene sequences. Nuncl Acids Res 32: W173-W175.
- Swelsen WTN, Voorter CEM, Van den Berg-Loonen EM (2002) Sequence analysis exons 1, 2, 3, 4 and 5 of the HLA-B5/35 cross-reacting group. Tissue Antigens 60: 224-234.
- Gao X, O’Brien TR, Welzel TM, Marti D, Qi Y, Goedert JJ, Phair J, Pfeiffer R, and Carrington M (2010) HLA-B alleles associate consistently with HIV heterosexual transmission, viral load and progression to AIDS, but not susceptibility to infection. AIDS 24: 1835-1840.
- Kilpatrick ED, Terajima M, Koster FT, Catalina MD, Cruz J, Ennis FA (2004) Role of specific CD8+ T cells in the severity of a fulminant zoonotic viral hemorrhagic fever, hantavirus pulmonary syndrome. J Immunol 172: 3297–3304.
- Marcos CY, Fernández-Viña MA, Lázaro AM, Nulf CJ, Raimondi EH, et al. (1997) Novel HLA-B35 subtypes: putative gene conversion events with donor sequences from alleles common in natives Americans (HLA-B*4002 or B*4801). Hum Immunol 53: 148-155.
- Ferrer JF, Jonsson CB, Esteban E, Galligan D, Basombrio MA, et al. (1998) High Prevalence of hantavirus infection in Indian communities of the Paraguayan and Argentinean Gran Chaco. Am J Trop Med Hyg 59: 438–444.
- Armien B, Pascale J, Bayard V, Muñoz C, Mosca I, et al. (2004) High seroprevalence of hantavirus infection on the Azuero peninsula of Panamá. Am J Trop Med Hyg 70: 682-687.
- Adams S, Barrachini K, Chen D, Robbins F, Wang L, et al. (2004) Ambiguous allele combinations in HLA Class I and class II sequence-based typing: when precise nucleotide sequencing leads to imprecise allele identification. J Transl Med 2: 30.
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, et al. (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329: 512-518.
- Gao X, Nelson G, Karacki P, Martin M, Phair J, et al. (2001) Effect of a single amino acid change in MHC class I molecules on the rate of progression to aids. N Engl J Med 344: 1668-1675.
- Tynan F, Elhassen D, Purcell A, Burrows J, Borg N, et al. (2005) The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J Exp Med 202: 1249-1260.
- Beck Y, Satz L, Takamiya Y, Nakayama S, Ling L (1995) Polymorphism of Human minor histocompatibility antigens: T cell recognition of human minor histocompatibility peptides presented by HLA-B35 subtype molecules. J Exp Med 181: 2037-2048.
- Kalergis A, Fierro A, Figueroa C, González P, Tobar J (2004) La detección antígeno-específica de linfocitos T y sus aplicaciones clínicas. Rev Méd Chile 132: 371-380.
- Manigold T, Mori A, Graumann R, Llop E, Simon V, et al. (2010) Highly Differentiated, Resting Gn-Specific Memory CD8+ T Cells Persist Years after Infection by Andes Hantavirus. PLoS Pathogens 6: e1000779.


